TB MAC seminar series: Calibrating complex models with emulation & history matching (Andy Iskauskas)
Seminar summary:
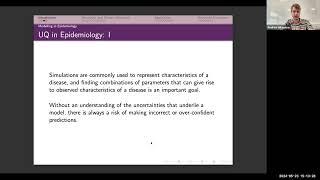
Complex models of disease transmission and dynamics are paramount in investigating the impact of interventions or making predictions of future outcomes. However, their complexity can be a barrier to an exhaustive exploration of parameter space, whether as a result of stochasticity, high-dimensional input space, or computational expense of simulator runs. To surmount this problem one may use the twin methodologies of emulation (a means by which a potentially slow simulator can be represented by a fast statistical surrogate) coupled with history matching (an exploration method that finds acceptable matches to observational data in the presence of uncertainty). Even where these techniques are well-defined, it may be difficult to implement such methods into a standard calibration workflow: I will demonstrate how the R package hmer enabled their application to a complex deterministic TB model across multiple countries, and discuss built-in extensions for stochastic simulators.
Presenter bio:
Andy Iskauskas is an Assistant Professor in Statistics and Willmore Fellow at Durham University, with particular focus on Bayesian calibration. He has worked with epidemiologists across the world modelling a number of different diseases, as well as non-epidemiological applications in physics, biology, and industry, and developed and maintains the history matching and emulation package hmer.
Complex models of disease transmission and dynamics are paramount in investigating the impact of interventions or making predictions of future outcomes. However, their complexity can be a barrier to an exhaustive exploration of parameter space, whether as a result of stochasticity, high-dimensional input space, or computational expense of simulator runs. To surmount this problem one may use the twin methodologies of emulation (a means by which a potentially slow simulator can be represented by a fast statistical surrogate) coupled with history matching (an exploration method that finds acceptable matches to observational data in the presence of uncertainty). Even where these techniques are well-defined, it may be difficult to implement such methods into a standard calibration workflow: I will demonstrate how the R package hmer enabled their application to a complex deterministic TB model across multiple countries, and discuss built-in extensions for stochastic simulators.
Presenter bio:
Andy Iskauskas is an Assistant Professor in Statistics and Willmore Fellow at Durham University, with particular focus on Bayesian calibration. He has worked with epidemiologists across the world modelling a number of different diseases, as well as non-epidemiological applications in physics, biology, and industry, and developed and maintains the history matching and emulation package hmer.
Комментарии:
The NBA2K MyCareer Story Makes No Sense
Ben Casalinho
Launge wala Border and Tanot Mata Mandir India Pakistan Border |Hamari Gadi Fas Gayi|Amit Rajput
Jivan ka safar with Amit Rajput
Прогулка в осенний день.
Моя жизнь в деревне
Where Bay Area Housing Market is Headed in 2025
Bay Area California Real Estate - Spencer Hsu
Это неизбежно… #реалитишоу #dubai #дубай #юмор #клининг #отношения
РЕАЛИТИ ШОУ ЧИСТО ДУБАЙ